Jevvy Huang describes the findings of a mini project that investigated whether a range of tools could provide useful insight into the carbon footprint of the computational processes in our research. (more…)
Author: IEU admin
Learning and adapting to make PPIE work better in non-clinical research
In a post based on a recent talk, Siân Harris shares some of what we have learned from the first four years of involving a patient advisory group in non-clinical cancer research in the Integrative Cancer Epidemiology Programme. (more…)
Adiposity in childhood affects the risk of breast cancer by changing breast tissue composition, study suggests

Breast cancer is the most common cancer in women worldwide. With rates continuing to rise, there is an urgent need to identify new modifiable breast cancer risk factors. New research led by the University of Bristol suggests that higher adiposity in childhood leads to less dense breast tissue forming, which results in a reduced breast cancer risk. However, further research is needed to understand the mechanism of the overall protective effect of childhood adiposity to identify new targets for intervention and prevention. (more…)
A novel measure of inflammation in depression
Éimear Foley discusses a recent paper
Inflammation is thought to play a role in depression. According to research that combined information from many studies (i.e., systematic review and meta-analysis), approximately a quarter of all those diagnosed with depression also have consistently higher levels of inflammatory proteins, like C-reactive protein (CRP), in their blood. Large population-based studies and studies using genetic information (i.e., Mendelian Randomization) further suggest that inflammation, particularly an inflammatory protein called interleukin 6 (IL-6), play a key role in causing depression. (You can read more about the role of inflammation in depression in our IEUREKA blog on “Immune cells as biomarkers of depression”.)
So far, studies examining this relationship between inflammation and depression have only focused on the levels of individual immune proteins in the blood, like IL-6 and CRP. At a cellular level, an inflammatory response is not caused by the activity of one inflammatory protein but rather by the interaction between several proteins working together to trigger a signalling response. Looking at the activity of proteins gives further insight into how these proteins relate to a particular condition, like depression. For this reason, in a recent study, we examined how IL-6 activity relates to symptoms and cognitive performance in people diagnosed with depression. (more…)
Environment and sustainability in health research – and what we are doing about it

In our research unit we are looking at ways to reduce the environmental impact of our research. Chin Yang Shapland and Hayley Wragg share some of what they learned in a recent workshop and how IEU is beginning to apply this learning in making our computing processes more sustainable. (more…)
Lessons from organising and participating in the University of Bristol Faculty of Health Sciences PGR Symposium
 PhD student Winfred Gatua was part of the team that organised a recent Postgraduate Research Symposium. In this post, she shares how organising this event helped her develop important skills for her research and beyond. (more…)
PhD student Winfred Gatua was part of the team that organised a recent Postgraduate Research Symposium. In this post, she shares how organising this event helped her develop important skills for her research and beyond. (more…)
Multi-ancestry genetic analysis of eczema highlights relevant pathways and potential drug targets
Ashley Budu-Aggrey and Lavinia Paternoster discuss their recent paper shedding light on genetic factors in eczema and the potential implications of understanding these links better. (more…)
Supporting Mendelian randomization in Uganda

In July 2023 a group of researchers from IEU travelled to Entebbe, Uganda to deliver a short course on genome-wide association analyses and Mendelian randomization. Amanda Chong shares a glimpse of what happened during the week.
EASD 2023: At the interface between diabetes research and clinical practice
PhD student Maddy Smith shares what she learned from attending a recent international conference on diabetes research.
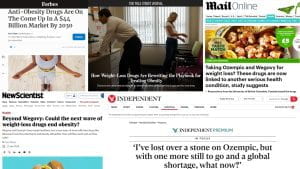
An exciting time for metabolic health
Weight loss drugs such as Wegovy and Ozempic have made many headlines and sparked a frenzy on social media in recent months. Not only is their potential to “end obesity” being unpacked in the media, but also their issues such as side effects, weight regain after stopping taking it, and supply shortages. Since obesity is a main risk factor for type 2 diabetes, these new drugs have implications for diabetes treatment – in fact, some of them were originally developed to treat diabetes and weight loss was noticed to be a concurrent effect.
Is a little bit of alcohol good for you?
Some things seem to be too good to be true.
The idea that life’s little pleasures – a glass of red wine, for example – might be good for us is seductive. Given that most public health advice is simultaneously common sense and mildly dispiriting – eat healthily, exercise, don’t smoke – the possibility that we don’t have to live like Spartans to live long and healthily lives is surely good news… But is that really the case?
In this blog post, first published on the Institute of Alcohol Studies blog, Professor Marcus Munafò and Professor George Davey Smith of MRC Integrative Epidemiology Unit explain why confounding factors skew studies to suggest a small amount of alcohol might have health benefits, and how Mendelian randomization has debunked this common myth. (more…)
