 Dr Jack Bowden, Programme Leader
Dr Jack Bowden, Programme Leader
Every two years my department puts on a conference on the topic of Mendelian Randomization (MR), a field that has been pioneered by researchers in Bristol over the last two decades. After months of planning, including finding a venue, inviting speakers from around the world and arranging the scientific programme, it’s a week and a half to go and we’re almost there!
But what is Mendelian Randomization research all about I hear you ask? Are you sure you want to know? Please read on but understand there is no going back…..

Have you ever had the feeling that something wasn’t quite right, that you are being controlled in some way by a higher force?
Well, it’s true. We are all in The Matrix. Like it or not, each of us has been recruited into an experiment from the moment we were born. Our genes, which are given to us by our parents at the point of conception, influence every aspect of our lives: how much we eat, sleep, drink, weigh, smoke, study, worry and play. The controlling effect is cleverly very small, and scientists only discovered the pattern by taking measurements across large populations, so as individuals we generally don’t notice. But the effect is real, very real!
How can we fight back?
We cannot escape The Matrix, but we can fight back by extracting knowledge from this unfortunate experiment we find ourselves in and using it for society’s advantage. For example, if we know that our genes predict 1-2% of variation in Low-Density Lipoprotein cholesterol (LDL-c – the ‘bad’ cholesterol) in the population, we can see if genes known to predict LDL-c also predict later life health outcomes in a group of individuals such as an increased risk of heart disease. If they do, then it provides strong evidence that reducing LDL-c will reduce heart disease risk, and we can then take steps to act. This is, in essence, the science of Mendelian randomization. See here for a nice animation of the method by our Unit director, George Davey Smith – our Neo if you like.

Mendelian randomization is very much a team effort, involving scientists with expertise across many disciplines. My role, as a statistician and data scientist is to provide the mathematical framework to ensure the analysis is performed in a rigorous and reliable manner.
We start by drawing a diagram that makes explicit the assumptions our analysis rests on. The arrows show which factors influence which. In our case we must assume that a set of genes influence LDL-c, and can only influence heart disease risk through LDL-c. We can then translate this diagram into a system of equations that we apply to our data.
The great thing about Mendelian randomization is that, even when many other factors jointly influence LDL-c and heart disease risk, the Mendelian randomization approach should still work.
Recently, the validity of the Mendelian randomization approach has been called into question due to the problem of pleiotropy. In our example this would be when a gene affects heart disease through a separate unmodelled pathway.
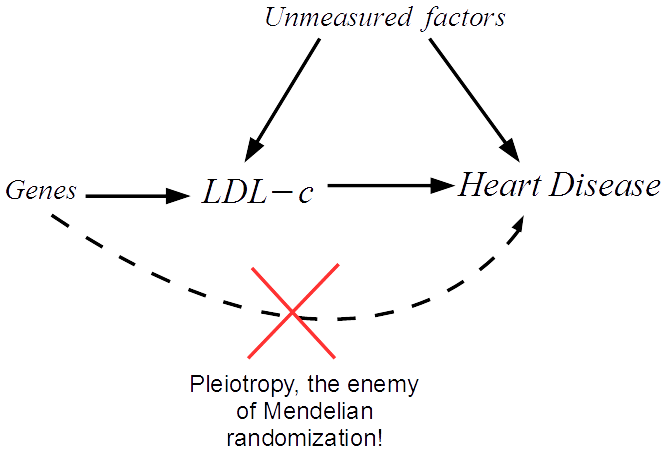
This can lead to bias in the analysis and therefore misleading results. My research is focused on novel methods that try to overcome the issue of pleiotropy, by detecting and adjusting for its presence in the analysis. For further details please see this video.
The MR Data challenge
At this year’s conference we are organising an MR Data Challenge, to engage conference participants in exploring and developing innovative approaches to Mendelian randomization using a publicly available data set. At a glance, the data comprises information on 150 genes and their association with
- 118 lipid measurements (LDL cholesterol)
- 7 health outcomes (including type II diabetes)
Eight research teams have submitted an entry to the competition, to describe how they would analyse the data and the conclusions they would draw. The great thing about these data is that the information on all 118 lipid traits simultaneously assessed to improve the robustness of the Mendelian randomization analysis.
A key aim of the session is to bring together data scientists with experts from the medical world to comment on and debate the results. We will publish all of the computer code online so that anyone can re-run the analyses. In the future, we hope to add further data to this resource and for many new teams to join the party with their own analysis attempt.
Please come and join us at the MR conference in Bristol, 17-19 July, it promises to be epic!

